Molecular Modelling
Understanding the Catalytic Mechanism of DMML
DMML is a member of the ICL superfamily which is widely distributed among microorganisms including bacteria such as Aspergillus. We report herein QM/MM studies aimed at improving our understanding of the catalytic events that take place within DMML. To this end we have generated density functional theory (DFT)/AMBER QM/MM models to explore the two main mechanistic proposals put forward on the basis of experimental structural data, while we also try to identify which are the most likely acidic and basic residues involved in each scenario. Shedding light on the mechanistic processes at work within this family may open up opportunities to modulate its activity.
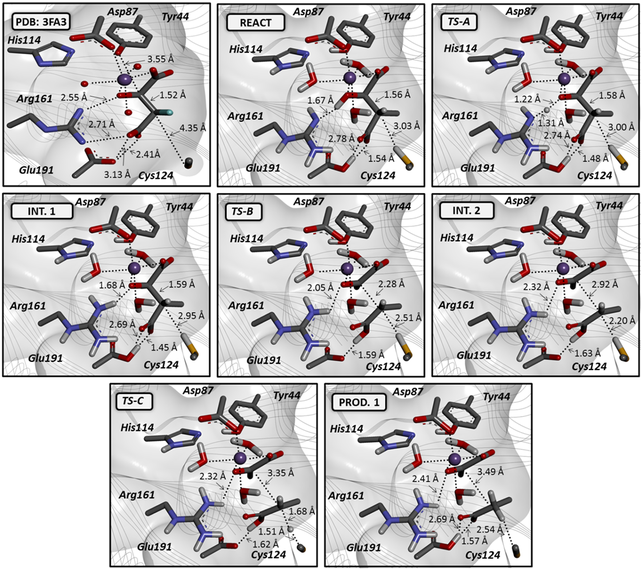
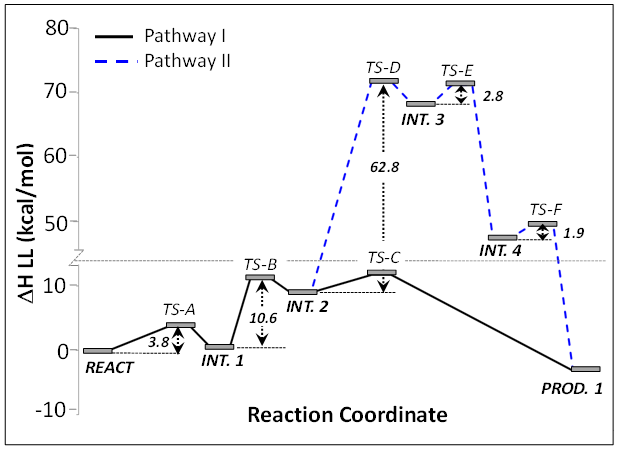
Our QM/MM calculations suggest that the most probable pathway involves three separate steps. In step 1, Arg161 acts as the general base that initiates the reaction by abstracting a proton from the C(2) hydroxyl of the substrate. Step 2 involves C(2)−C(3) bond cleavage, which is preceded by proton transfer from Glu191 to the C(4) carboxyl group. This is the ratedetermining step with a predicted barrier of approximately 9−10 kcal/mol. In step 3, Cys124 was identified as the general acid leading to the formation of propionate.
Our QM/MM calculations suggest that the most probable pathway involves three separate steps. In step 1, Arg161 acts as the general base that initiates the reaction by abstracting a proton from the C(2) hydroxyl of the substrate. Step 2 involves C(2)−C(3) bond cleavage, which is preceded by proton transfer from Glu191 to the C(4) carboxyl group. This is the ratedetermining step with a predicted barrier of approximately 9−10 kcal/mol. In step 3, Cys124 was identified as the general acid leading to the formation of propionate.
Article weblink: http://pubs.acs.org/doi/10.1021/acs.jpcb.5b04732
Article weblink: https://doi.org/10.1016/j.jmgm.2016.05.010
Article weblink: https://doi.org/10.1016/j.jmgm.2016.05.010
|
| ||||||||||||