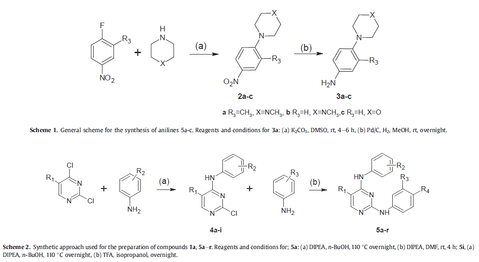
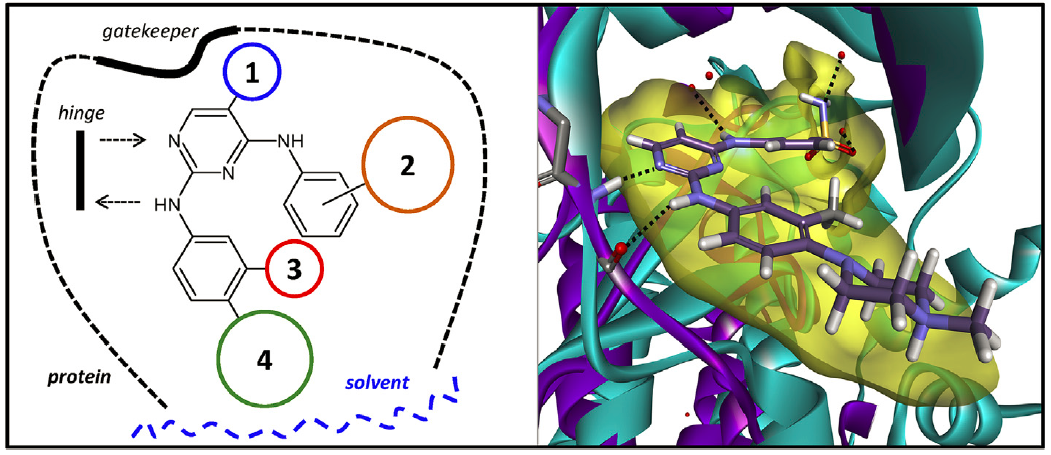
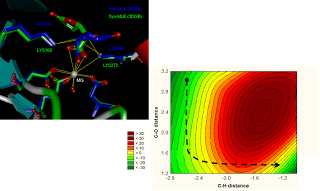
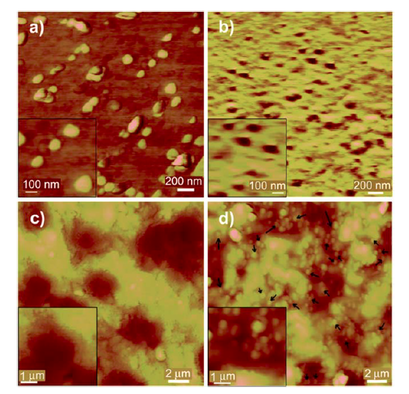
The Computational and Medicinal Chemistry Laboratory (CMCL) is located within the Department of Biomedical Engineering, Faculty of Engineering, King Mongkut's Institute of Technology Ladkrabang. Our research interests lie in the Pharmaceutical Engineering field, a multi-disciplinary area that encompasses the fields of Chemistry, Physics, Biochemistry, Biology, Engineering and Medicine.
There is an opening for a fully funded Ph.D. position: Click here for more details (pdf).
There is an opening for a fully funded Ph.D. position: Click here for more details (pdf).